Enzyme found in humans
| LIG4 |
|---|
 |
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
| List of PDB id codes |
|---|
1IK9, 2E2W, 3II6, 3VNN, 3W1B, 3W1G, 3W5O, 4HTO, 4HTP |
|
|
| Identifiers |
|---|
| Aliases | LIG4, LIG4S, DNA ligase 4 |
|---|
| External IDs | OMIM: 601837; MGI: 1335098; HomoloGene: 1736; GeneCards: LIG4; OMA:LIG4 - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 13 (human)[1] |
|---|
| | Band | 13q33.3 | Start | 108,207,439 bp[1] |
|---|
| End | 108,218,368 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 8 (mouse)[2] |
|---|
| | Band | 8|8 A1.1 | Start | 10,019,049 bp[2] |
|---|
| End | 10,027,686 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - endothelial cell
- oocyte
- secondary oocyte
- mucosa of paranasal sinus
- retinal pigment epithelium
- Brodmann area 23
- islet of Langerhans
- jejunal mucosa
- primary visual cortex
- lower lobe of lung
|
| | Top expressed in | - primary oocyte
- secondary oocyte
- thymus
- zygote
- hand
- trigeminal ganglion
- otolith organ
- epithelium of lens
- utricle
- substantia nigra
|
| | More reference expression data |
|
|---|
| BioGPS |  | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - nucleotide binding
- metal ion binding
- protein C-terminus binding
- protein binding
- ATP binding
- DNA ligase (ATP) activity
- ligase activity
- DNA binding
- DNA ligase activity
| | Cellular component | - nucleoplasm
- DNA ligase IV complex
- nonhomologous end joining complex
- condensed chromosome
- nucleus
- DNA-dependent protein kinase-DNA ligase 4 complex
- cytoplasm
- cytoplasmic ribonucleoprotein granule
| | Biological process | - nucleotide-excision repair, DNA gap filling
- double-strand break repair via classical nonhomologous end joining
- response to ionizing radiation
- DNA recombination
- lagging strand elongation
- DNA biosynthetic process
- cellular response to lithium ion
- single strand break repair
- cellular response to DNA damage stimulus
- cell division
- DNA replication
- positive regulation of chromosome organization
- establishment of integrated proviral latency
- cell cycle
- response to X-ray
- positive regulation of neurogenesis
- DNA ligation involved in DNA recombination
- DNA ligation
- DNA ligation involved in DNA repair
- somatic stem cell population maintenance
- chromosome organization
- T cell receptor V(D)J recombination
- neuron apoptotic process
- isotype switching
- T cell differentiation in thymus
- central nervous system development
- immunoglobulin V(D)J recombination
- double-strand break repair via nonhomologous end joining
- positive regulation of fibroblast proliferation
- in utero embryonic development
- cell population proliferation
- V(D)J recombination
- pro-B cell differentiation
- DNA repair
- response to gamma radiation
- double-strand break repair
- negative regulation of neuron apoptotic process
- cellular response to ionizing radiation
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | NM_001098268
NM_002312
NM_206937
NM_001330595
NM_001352598
|
|---|
NM_001352599
NM_001352600
NM_001352601
NM_001352602
NM_001352603
NM_001352604
NM_001379095 |
| |
|---|
| RefSeq (protein) | NP_001091738
NP_001317524
NP_002303
NP_996820
NP_001339527
|
|---|
NP_001339528
NP_001339529
NP_001339530
NP_001339531
NP_001339532
NP_001339533
NP_001366024 |
| |
|---|
| Location (UCSC) | Chr 13: 108.21 – 108.22 Mb | Chr 8: 10.02 – 10.03 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
DNA ligase 4 also DNA ligase IV, is an enzyme that in humans is encoded by the LIG4 gene.[5]
Function
DNA ligase 4 is an ATP-dependent DNA ligase that joins double-strand breaks during the non-homologous end joining pathway of double-strand break repair. It is also essential for V(D)J recombination. Lig4 forms a complex with XRCC4, and further interacts with the DNA-dependent protein kinase (DNA-PK) and XLF/Cernunnos, which are also required for NHEJ. The crystal structure of the Lig4/XRCC4 complex has been resolved.[6] Defects in this gene are the cause of LIG4 syndrome. The yeast homolog of Lig4 is Dnl4.
LIG4 syndrome
In humans, deficiency of DNA ligase 4 results in a clinical condition known as LIG4 syndrome. This syndrome is characterized by cellular radiation sensitivity, growth retardation, developmental delay, microcephaly, facial dysmorphisms, increased disposition to leukemia, variable degrees of immunodeficiency and reduced number of blood cells.[7][8]
Haematopoietic stem cell aging
Accumulation of DNA damage leading to stem cell exhaustion is regarded as an important aspect of aging.[9][10] Deficiency of lig4 in pluripotent stem cells impairs Non-homologous end joining (NHEJ) and results in accumulation of DNA double-strand breaks and enhanced apoptosis.[8] Lig4 deficiency in the mouse causes a progressive loss of haematopoietic stem cells and bone marrow cellularity during aging.[11] The sensitivity of haematopoietic stem cells to lig4 deficiency suggests that lig4-mediated NHEJ is a key determinant of the ability of stem cells to maintain themselves against physiological stress over time.[8][11]
Interactions
LIG4 has been shown to interact with XRCC4 via its BRCT domain.[12][6] This interaction stabilizes LIG4 protein in cells; cells that are deficient for XRCC4, such as XR-1 cells, have reduced levels of LIG4.[13]
Mechanism
LIG4 is an ATP-dependent DNA ligase. LIG4 uses ATP to adenylate itself and then transfers the AMP group to the 5' phosphate of one DNA end. Nucleophilic attack by the 3' hydroxyl group of a second DNA end and release of AMP yield the ligation product. Adenylation of LIG4 is stimulated by XRCC4 and XLF.[14]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000174405 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000049717 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: LIG4 ligase IV, DNA, ATP-dependent".
- ^ a b Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L (December 2001). "Crystal structure of an Xrcc4-DNA ligase IV complex". Nature Structural Biology. 8 (12): 1015–9. doi:10.1038/nsb725. PMID 11702069. S2CID 21218268.
- ^ Rucci F, Notarangelo LD, Fazeli A, Patrizi L, Hickernell T, Paganini T, Coakley KM, Detre C, Keszei M, Walter JE, Feldman L, Cheng HL, Poliani PL, Wang JH, Balter BB, Recher M, Andersson EM, Zha S, Giliani S, Terhorst C, Alt FW, Yan CT (February 2010). "Homozygous DNA ligase IV R278H mutation in mice leads to leaky SCID and represents a model for human LIG4 syndrome". Proceedings of the National Academy of Sciences of the United States of America. 107 (7): 3024–9. Bibcode:2010PNAS..107.3024R. doi:10.1073/pnas.0914865107. PMC 2840307. PMID 20133615.
- ^ a b c Tilgner K, Neganova I, Moreno-Gimeno I, Al-Aama JY, Burks D, Yung S, Singhapol C, Saretzki G, Evans J, Gorbunova V, Gennery A, Przyborski S, Stojkovic M, Armstrong L, Jeggo P, Lako M (August 2013). "A human iPSC model of Ligase IV deficiency reveals an important role for NHEJ-mediated-DSB repair in the survival and genomic stability of induced pluripotent stem cells and emerging haematopoietic progenitors". Cell Death and Differentiation. 20 (8): 1089–100. doi:10.1038/cdd.2013.44. PMC 3705601. PMID 23722522.
- ^ Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL (June 2007). "Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age". Nature. 447 (7145): 725–9. Bibcode:2007Natur.447..725R. doi:10.1038/nature05862. PMID 17554309. S2CID 4416445.
- ^ Bernstein H, Payne CM, Bernstein C, Garewal H, Dvorak K (2008). "Chapter 1: Cancer and aging as consequences of un-repaired DNA damage". In Kimura H, Suzuki A (eds.). New Research on DNA Damages. New York: Nova Science Publishers, Inc. pp. 1–47. ISBN 978-1-60456-581-2. Archived from the original on 2014-10-25. Retrieved 2015-05-20.
- ^ a b Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ (June 2007). "DNA repair is limiting for haematopoietic stem cells during ageing". Nature. 447 (7145): 686–90. Bibcode:2007Natur.447..686N. doi:10.1038/nature05875. PMID 17554302. S2CID 4332976.
- ^ Deshpande RA, Wilson TE (October 2007). "Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4". DNA Repair. 6 (10): 1507–16. doi:10.1016/j.dnarep.2007.04.014. PMC 2064958. PMID 17567543.
- ^ Bryans M, Valenzano MC, Stamato TD (January 1999). "Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4". Mutation Research. 433 (1): 53–8. doi:10.1016/s0921-8777(98)00063-9. PMID 10047779.
- ^ Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP (February 2013). "XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair". Biochemistry and Cell Biology. 91 (1): 31–41. doi:10.1139/bcb-2012-0058. PMC 3725335. PMID 23442139.
Further reading
- Wei YF, Robins P, Carter K, Caldecott K, Pappin DJ, Yu GL, Wang RP, Shell BK, Nash RA, Schär P (June 1995). "Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination". Molecular and Cellular Biology. 15 (6): 3206–16. doi:10.1128/mcb.15.6.3206. PMC 230553. PMID 7760816.
- Robins P, Lindahl T (September 1996). "DNA ligase IV from HeLa cell nuclei". The Journal of Biological Chemistry. 271 (39): 24257–61. doi:10.1074/jbc.271.39.24257. PMID 8798671.
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR (July 1997). "Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells". Nature. 388 (6641): 492–5. doi:10.1038/41358. PMID 9242410. S2CID 4349909.
- Critchlow SE, Bowater RP, Jackson SP (August 1997). "Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV". Current Biology. 7 (8): 588–98. Bibcode:1997CBio....7..588C. doi:10.1016/S0960-9822(06)00258-2. PMID 9259561.
- Grawunder U, Zimmer D, Lieber MR (July 1998). "DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains". Current Biology. 8 (15): 873–6. Bibcode:1998CBio....8..873G. doi:10.1016/S0960-9822(07)00349-1. PMID 9705934.
- Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR (October 1998). "DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes". Molecular Cell. 2 (4): 477–84. doi:10.1016/S1097-2765(00)80147-1. PMID 9809069.
- Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, Lehmann AR, Jackson SP, Jeggo PA (July 1999). "Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient". Current Biology. 9 (13): 699–702. Bibcode:1999CBio....9..699R. doi:10.1016/S0960-9822(99)80311-X. PMID 10395545.
- Kim ST, Lim DS, Canman CE, Kastan MB (December 1999). "Substrate specificities and identification of putative substrates of ATM kinase family members". The Journal of Biological Chemistry. 274 (53): 37538–43. doi:10.1074/jbc.274.53.37538. PMID 10608806.
- Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA (May 2000). "Ku recruits the XRCC4-ligase IV complex to DNA ends". Molecular and Cellular Biology. 20 (9): 2996–3003. doi:10.1128/MCB.20.9.2996-3003.2000. PMC 85565. PMID 10757784.
- Chen L, Trujillo K, Sung P, Tomkinson AE (August 2000). "Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase". The Journal of Biological Chemistry. 275 (34): 26196–205. doi:10.1074/jbc.M000491200. PMID 10854421.
- Lee KJ, Huang J, Takeda Y, Dynan WS (November 2000). "DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system". The Journal of Biological Chemistry. 275 (44): 34787–96. doi:10.1074/jbc.M004011200. PMID 10945980.
- Riballo E, Doherty AJ, Dai Y, Stiff T, Oettinger MA, Jeggo PA, Kysela B (August 2001). "Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity". The Journal of Biological Chemistry. 276 (33): 31124–32. doi:10.1074/jbc.M103866200. PMID 11349135.
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L (December 2001). "Crystal structure of an Xrcc4-DNA ligase IV complex". Nature Structural Biology. 8 (12): 1015–9. doi:10.1038/nsb725. PMID 11702069. S2CID 21218268.
- O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, Gatti RA, Varon R, Oettinger MA, Neitzel H, Jeggo PA, Concannon P (December 2001). "DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency". Molecular Cell. 8 (6): 1175–85. doi:10.1016/S1097-2765(01)00408-7. PMID 11779494.
- Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD, Dunning A (June 2002). "Variants in DNA double-strand break repair genes and breast cancer susceptibility". Human Molecular Genetics. 11 (12): 1399–407. doi:10.1093/hmg/11.12.1399. PMID 12023982.
- Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA (July 2002). "Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair". Molecular and Cellular Biology. 22 (14): 5194–202. doi:10.1128/MCB.22.14.5194-5202.2002. PMC 139779. PMID 12077346.
- Roth DB (July 2002). "Amplifying mechanisms of lymphomagenesis". Molecular Cell. 10 (1): 1–2. doi:10.1016/S1097-2765(02)00573-7. PMID 12150897.
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (October 2002). "DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2" (PDF). Current Biology. 12 (19): 1635–44. Bibcode:2002CBio...12.1635S. doi:10.1016/S0960-9822(02)01179-X. PMID 12361565. S2CID 17749937. Archived from the original (PDF) on 2021-05-08. Retrieved 2019-08-19.
- Roddam PL, Rollinson S, O'Driscoll M, Jeggo PA, Jack A, Morgan GJ (December 2002). "Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination". Journal of Medical Genetics. 39 (12): 900–5. doi:10.1136/jmg.39.12.900. PMC 1757220. PMID 12471202.

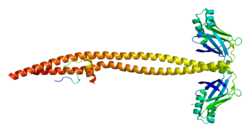
 1ik9: CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX
1ik9: CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX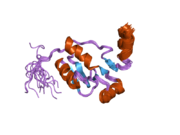 2e2w: Solution structure of the first BRCT domain of human DNA ligase IV
2e2w: Solution structure of the first BRCT domain of human DNA ligase IV



















